Instructions of using ISGPRE
ISGPRE is constructed based on the Genome-build GRCh38.p12.
It provides high-throughput predictions for putative interferon stimulated human genes.
Features used to build our prediction model can be found HERE.
Some of results are supported with experimental evidences from OCISG or Interferome databases.
You can quickly make your own predictions by following a few steps.
For more information please contact us via: david.l.robertson@glasgow.ac.uk, joseph.hughes@glasgow.ac.uk or h.chai.research@gmail.com
STEP ONE: SUBMIT YOUR IDENTIFIERS
You do not need to input or upload your genetic sequences. The only thing ISGPRE wants is the identifier of your human genes.
ISGPRE accepts FIVE types of identifiers including: HGNC ID, RefSeqAcc, NCBI gene ID, Ensembl gene ID, and Approved Symbol.
You can input different types of identifiers or unify them with the matching service from UniProt.
ISGPRE does not set restrictions on the number of your identifiers, but they should be valid in the right format and be separated by commas or newlines.
Here, we take ISG: IFITM3, MX1 and HPSE as examples to introduce how this webserver works.

In this example, Q1 is a gene identifier used by HUGO Gene Nomenclature Committee. Please avoid using numeric ID directly (e.g. Q6) when inputting these identifiers.
Q2 is a nucleotide identifier used by NCBI Reference Sequence Database.
Q3 is a gene identifier used by Ensembl.
Q4 is a gene identifier used by National Center for Biotechnology Information. Please avoid using numeric ID directly (e.g. Q6) when inputting these identifiers.
Q5 is an approved symbol for a specific gene or protein. ISGPRE provides case sensitive matching for approved symbols. Thus do not input symbols like Q7.
STEP TWO: REVIEW THE RESULTS
You can remove all you inputs by click the CLEAR button or get the results of your prediction soon after you click the PREDICT button on the right:
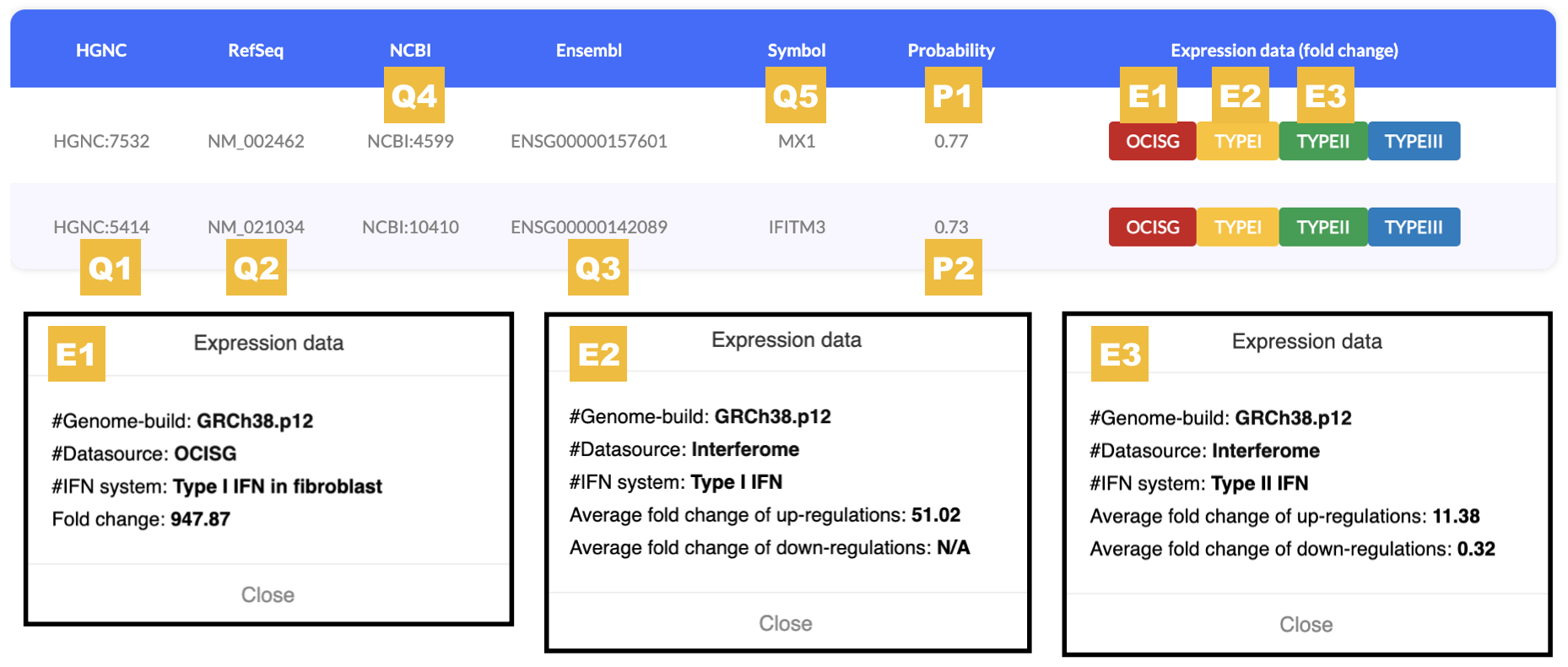
In this example, we get 2 results based on the input 7 identifiers.
Q1, Q2 and Q3 are identifiers for IFITM3; Q4 and Q5 are identifiers for MX1.
Results about HPSE are not shown as the input identifiers, i.e., Q6: 5164 (should be 'HGNC:5164') or Q7: hpse (should be 'HPSE') are not valid.
P1 and P2 are prediction scores for IFITM3 and MX1, respectively.
This prediction score can be used to measure the probability of being ISGs for your genes of interest.
In general, higher score indicates higher possibility of being a ISG.
We recommend to use 0.549 as a threshold to judge the prediction scores.
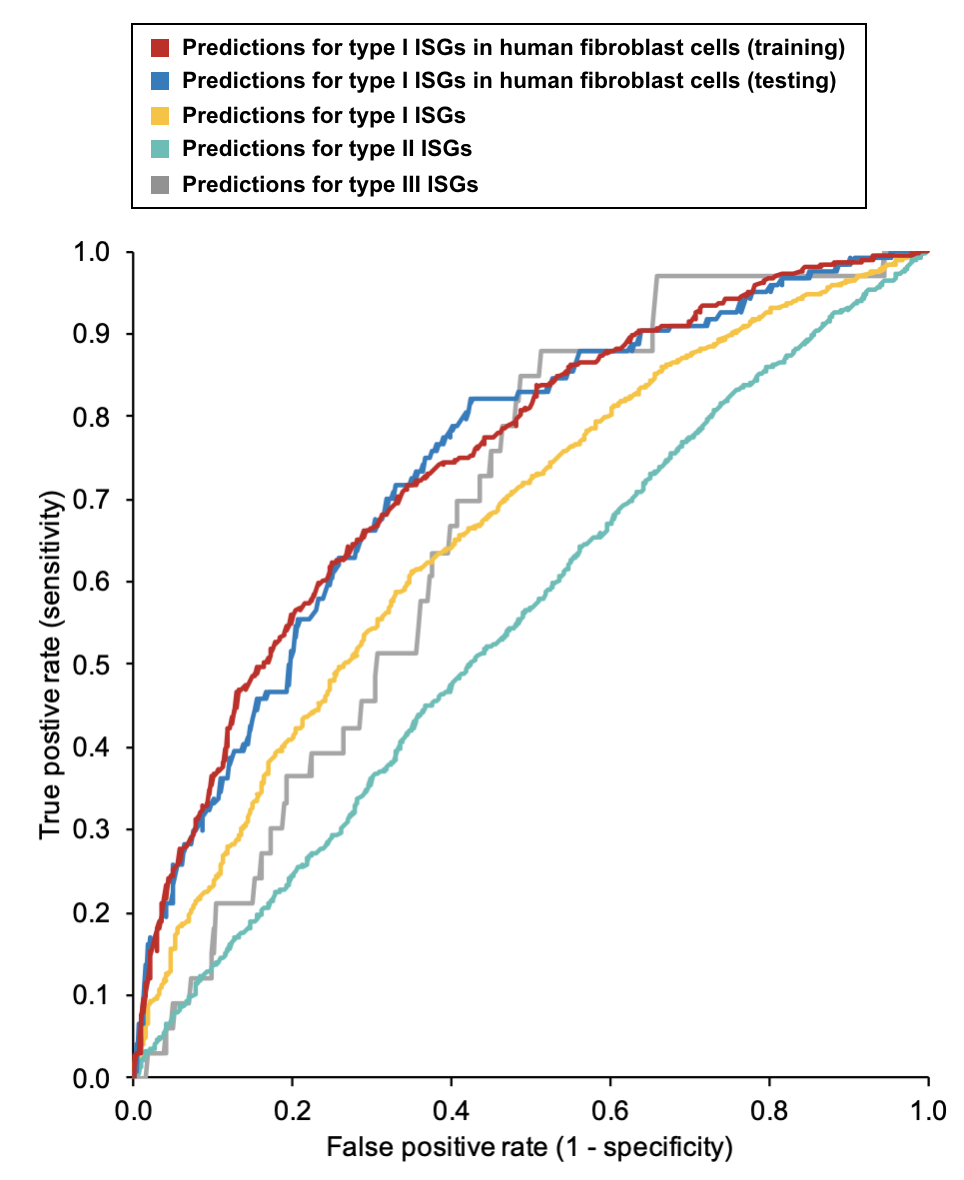
It should be noted that the prediction score works well to identify putative type I ISG in human fibroblast cells (see red and blue lines).
The prediction score can also be applied to discover putative new type I ISGs (see the yellow line), especially when there are no experimental data shown in the last column.
Due to functional similarity of type I and type III ISGs, the prediction scores can be referred when trying to identify type III ISGs (see the grey line).
The prediction score needs to be treated with caution for type II ISGs.
E1, E2 and E3 are experimental data retrieved from OCISG and Interferome databases.
These expression data are not annotated to all human genes, especially not to those with limited expressions in specific biological environment.